Compare {sdmgamindex} to regular GAMs
March 11 2024
C-model-comparisons.RmdGAMs: the old fashioned way
1. Wrangle Data
SPECIES <- c(# must match common name column
"walleye pollock",
"yellowfin sole",
"red king crab"
)
YEARS <- 2015:2021
SRVY <- "EBS"
method <- "ML"
knotsP <- 376
# dir_out <- here::here("inst","regular_gam_approach")
crs_latlon = "+proj=longlat +datum=WGS84"
crs_proj = "EPSG:3338"
dat <- sdmgamindex::noaa_afsc_public_foss
# Get cold pool data using cold pool data from the {coldpool} R package
dat <- dat %>%
dplyr::mutate(sx = ((longitude_dd_start - mean(longitude_dd_start, na.rm = TRUE))/1000),
sy = ((latitude_dd_start - mean(latitude_dd_start, na.rm = TRUE))/1000)) %>%
stats::na.omit() %>%
dplyr::filter(common_name %in% SPECIES &
year %in% YEARS &
srvy %in% SRVY) %>%
dplyr::rename(GEAR_TEMPERATURE = bottom_temperature_c,
BOTTOM_DEPTH = depth_m,
Year = year)
head(dat)
#> Year srvy survey survey_definition_id cruise cruisejoin haul
#> 1 2021 EBS eastern Bering Sea 98 202101 -751 88
#> 2 2021 EBS eastern Bering Sea 98 202101 -752 59
#> 3 2016 EBS eastern Bering Sea 98 201601 -706 48
#> 4 2019 EBS eastern Bering Sea 98 201901 -727 28
#> 5 2018 EBS eastern Bering Sea 98 201801 -722 22
#> 6 2021 EBS eastern Bering Sea 98 202101 -751 19
#> hauljoin stratum station vessel_id vessel_name date_time
#> 1 -19996 20 L-01 94 VESTERAALEN 2021-06-21 15:21:16
#> 2 -19973 31 I-03 162 ALASKA KNIGHT 2021-06-14 12:10:16
#> 3 -15350 10 L-04 94 VESTERAALEN 2016-06-12 07:56:25
#> 4 -18780 31 B-08 162 ALASKA KNIGHT 2019-06-09 06:27:08
#> 5 -17559 31 I-12 94 VESTERAALEN 2018-06-06 12:32:44
#> 6 -19886 10 I-11 94 VESTERAALEN 2021-06-03 13:11:46
#> latitude_dd_start longitude_dd_start latitude_dd_end longitude_dd_end
#> 1 58.65480 -167.8979 58.67117 -167.8579
#> 2 57.66072 -166.4836 57.66197 -166.5305
#> 3 58.64714 -165.9241 58.67214 -165.9363
#> 4 55.34481 -163.3904 55.34473 -163.4362
#> 5 57.65918 -160.9100 57.68333 -160.9030
#> 6 57.69411 -161.5022 57.66878 -161.5077
#> GEAR_TEMPERATURE surface_temperature_c BOTTOM_DEPTH species_code itis worms
#> 1 3.7 6.8 47 69322 97935 233889
#> 2 3.2 5.7 67 69322 97935 233889
#> 3 6.5 6.8 37 69322 97935 233889
#> 4 5.4 8.7 53 69322 97935 233889
#> 5 4.1 6.1 56 69322 97935 233889
#> 6 4.1 5.7 53 69322 97935 233889
#> common_name scientific_name id_rank cpue_kgkm2 cpue_nokm2 count
#> 1 red king crab Paralithodes camtschaticus species 73.81732 43.11760 2
#> 2 red king crab Paralithodes camtschaticus species 69.57943 21.41565 1
#> 3 red king crab Paralithodes camtschaticus species 66.83110 70.34853 3
#> 4 red king crab Paralithodes camtschaticus species 127.86046 82.09339 4
#> 5 red king crab Paralithodes camtschaticus species 531.50033 274.20481 12
#> 6 red king crab Paralithodes camtschaticus species 1166.16772 964.34424 42
#> weight_kg taxon_confidence area_swept_km2 distance_fished_km duration_hr
#> 1 3.424 1 0.046385 2.952 0.535
#> 2 3.249 1 0.046695 2.805 0.519
#> 3 2.850 1 0.042645 2.869 0.524
#> 4 6.230 1 0.048725 2.914 0.523
#> 5 23.260 1 0.043763 2.716 0.498
#> 6 50.790 1 0.043553 2.834 0.512
#> net_width_m net_height_m performance sx sy
#> 1 15.713 2.299 0.0 0.0008104423 0.0004173062
#> 2 16.647 2.400 0.0 0.0022246523 -0.0005767738
#> 3 14.864 2.250 0.0 0.0027842223 0.0004096462
#> 4 16.721 2.244 6.3 0.0053179323 -0.0028926838
#> 5 16.113 3.144 0.0 0.0077982823 -0.0005783138
#> 6 15.368 2.572 0.0 0.0072060923 -0.00054338382. Formulas
fm <- list(
# Null model spatial and temporal with an additional year effect
"fm_1_s_t_st" = "Year +
s(sx,sy,bs=c('ts'),k=376) +
s(sx,sy,bs=c('ts'),k=10,by=Year)",
# Mdoel with simple covariates
"fm_2_cov" =
"s(BOTTOM_DEPTH,bs='ts',k=10) +
s(log(GEAR_TEMPERATURE+3),bs='ts',k=10)"
)3. Run simple GAM models
Here are all of the models we want to try fitting:
comb <- tidyr::crossing(
"SPECIES" = SPECIES,
"fm_name" = gsub(pattern = " ", replacement = "_", x = names(fm))) %>%
dplyr::left_join(
x = .,
y = data.frame("fm" = gsub(pattern = "\n", replacement = "",
x = unlist(fm), fixed = TRUE),
"fm_name" = gsub(pattern = " ", replacement = "_",
x = names(fm))),
by = "fm_name")
comb
#> # A tibble: 6 × 3
#> SPECIES fm_name fm
#> <chr> <chr> <chr>
#> 1 red king crab fm_1_s_t_st Year + s(sx,sy,bs=c('ts'),k=376) + s(sx,sy…
#> 2 red king crab fm_2_cov s(BOTTOM_DEPTH,bs='ts',k=10) +s(log(GEAR_TEMPERAT…
#> 3 walleye pollock fm_1_s_t_st Year + s(sx,sy,bs=c('ts'),k=376) + s(sx,sy…
#> 4 walleye pollock fm_2_cov s(BOTTOM_DEPTH,bs='ts',k=10) +s(log(GEAR_TEMPERAT…
#> 5 yellowfin sole fm_1_s_t_st Year + s(sx,sy,bs=c('ts'),k=376) + s(sx,sy…
#> 6 yellowfin sole fm_2_cov s(BOTTOM_DEPTH,bs='ts',k=10) +s(log(GEAR_TEMPERAT…
models <- fittimes <- list()
for(i in 1:nrow(comb)){
cat("Fitting ",comb$SPECIES[i],"\n", comb$fm_name[i], ": ", comb$fm[i], "\n")
temp <- paste0(comb$SPECIES[i], " ", comb$fm_name[i])
dat0 <- dat %>%
dplyr::filter(common_name %in% comb$SPECIES[i])
fittimes[[ temp ]] <-
system.time ( models[[ temp ]] <-
gam(formula = as.formula(paste0("cpue_kgha ~ ", comb$fm[i])),
data = dat0,
family = tw,
gamma = 1.4) )
}
save(models, fittimes, file = here::here("inst","VigC_model_output.rdata"))4. Assess the model
b <- lapply(X = models, FUN = AIC)
bb <- sapply(models, `[`, 1)
# sdmgamindex::get_surveyidx_aic(models)
b <- AIC(models$`yellowfin sole fm_1_s_t_st`, models$`yellowfin sole fm_2_cov`)# get(paste0("b", 1:9))
b
#> df AIC
#> models$`yellowfin sole fm_1_s_t_st` 105.06470 13881.31
#> models$`yellowfin sole fm_2_cov` 15.22028 14558.49
# bb <- get(rownames(b)[b$AIC %in% min(b$AIC):(min(b$AIC)+5)])
# b$df %in% min(b$df):(min(b$df)+5)
# bb
bb <- models$`yellowfin sole fm_1_s_t_st`
# gam.check(bb)
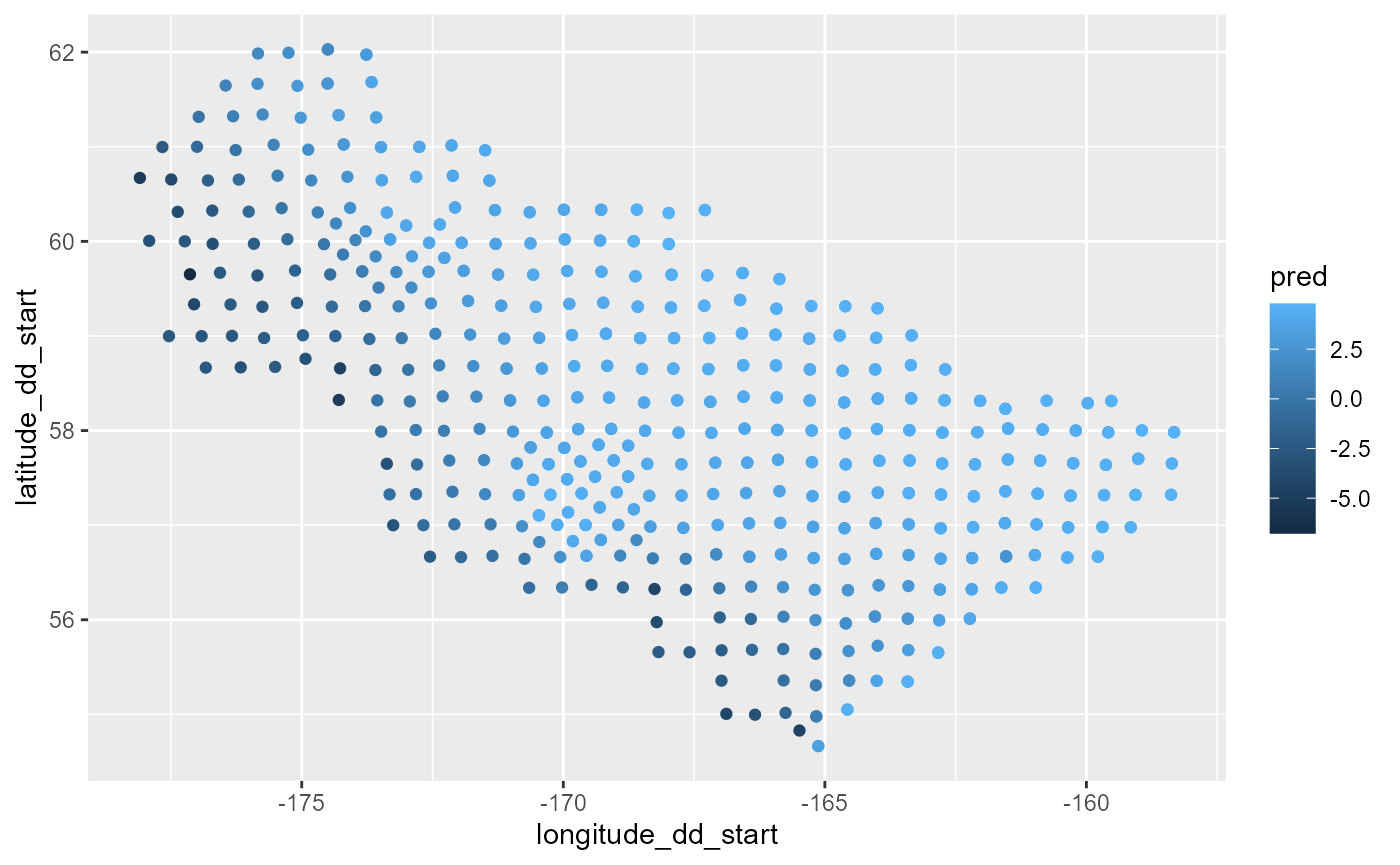
vis.gam(bb)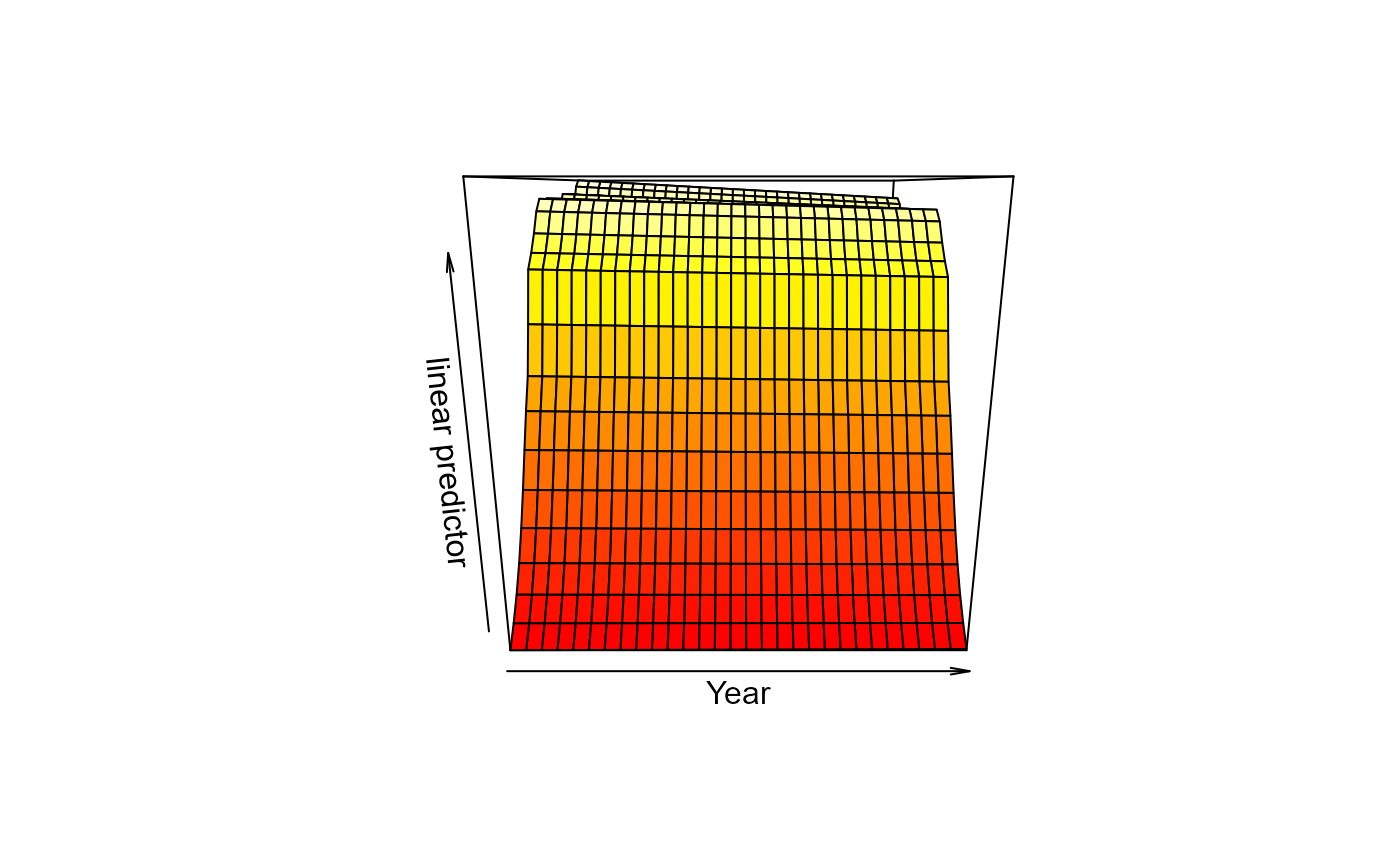
cc <- predict.gam(object = bb, newdata = dat[dat$Year == 2021,])
dat0 <- dat[dat$Year == 2021,] %>%
dplyr::mutate(pred = cc)
figure <- ggplot() +
geom_point(data = dat0,
mapping = aes(x= longitude_dd_start, y = latitude_dd_start, color = pred))
figure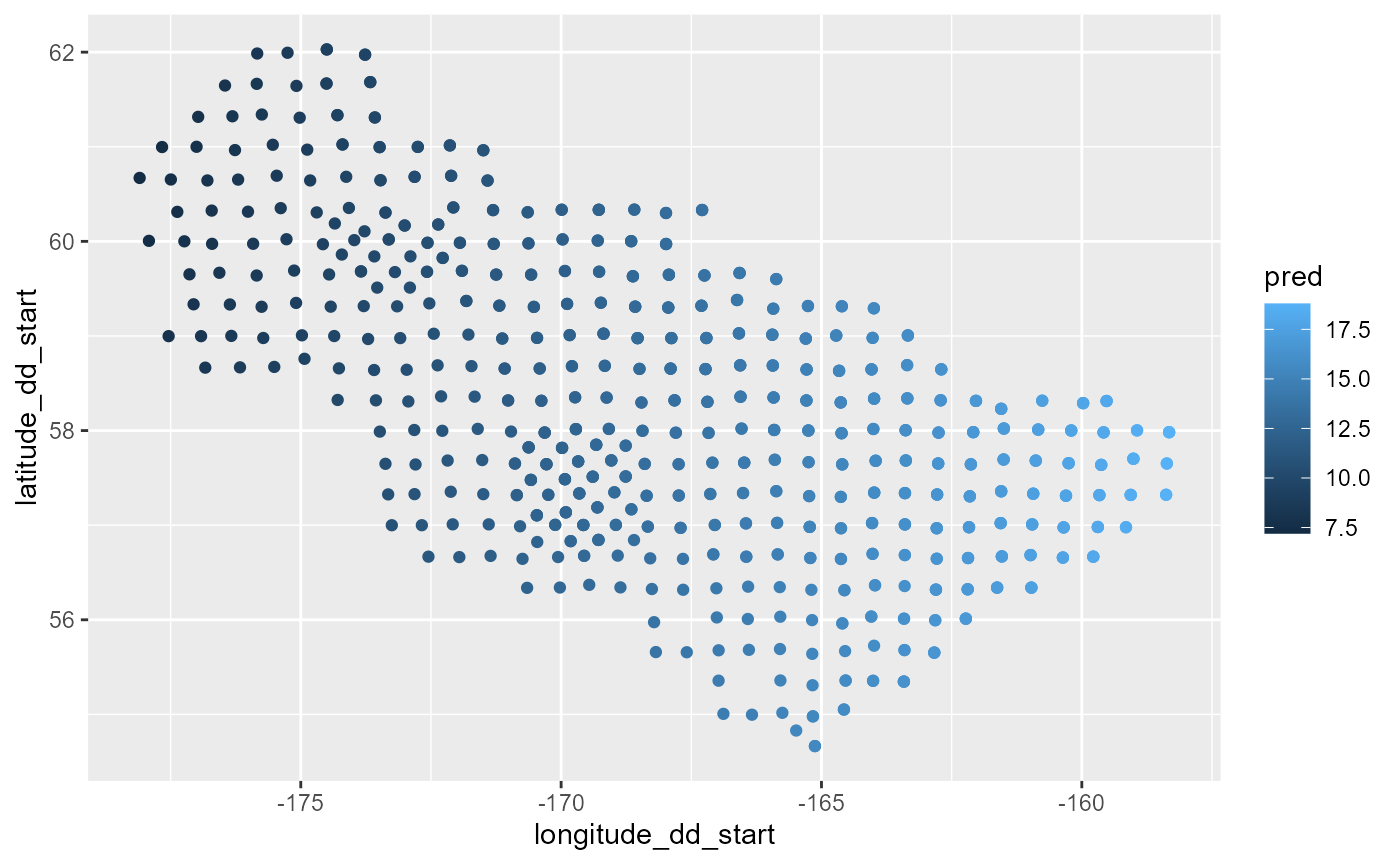
bb <- models$`yellowfin sole fm_2_cov`
# gam.check(bb)
vis.gam(bb)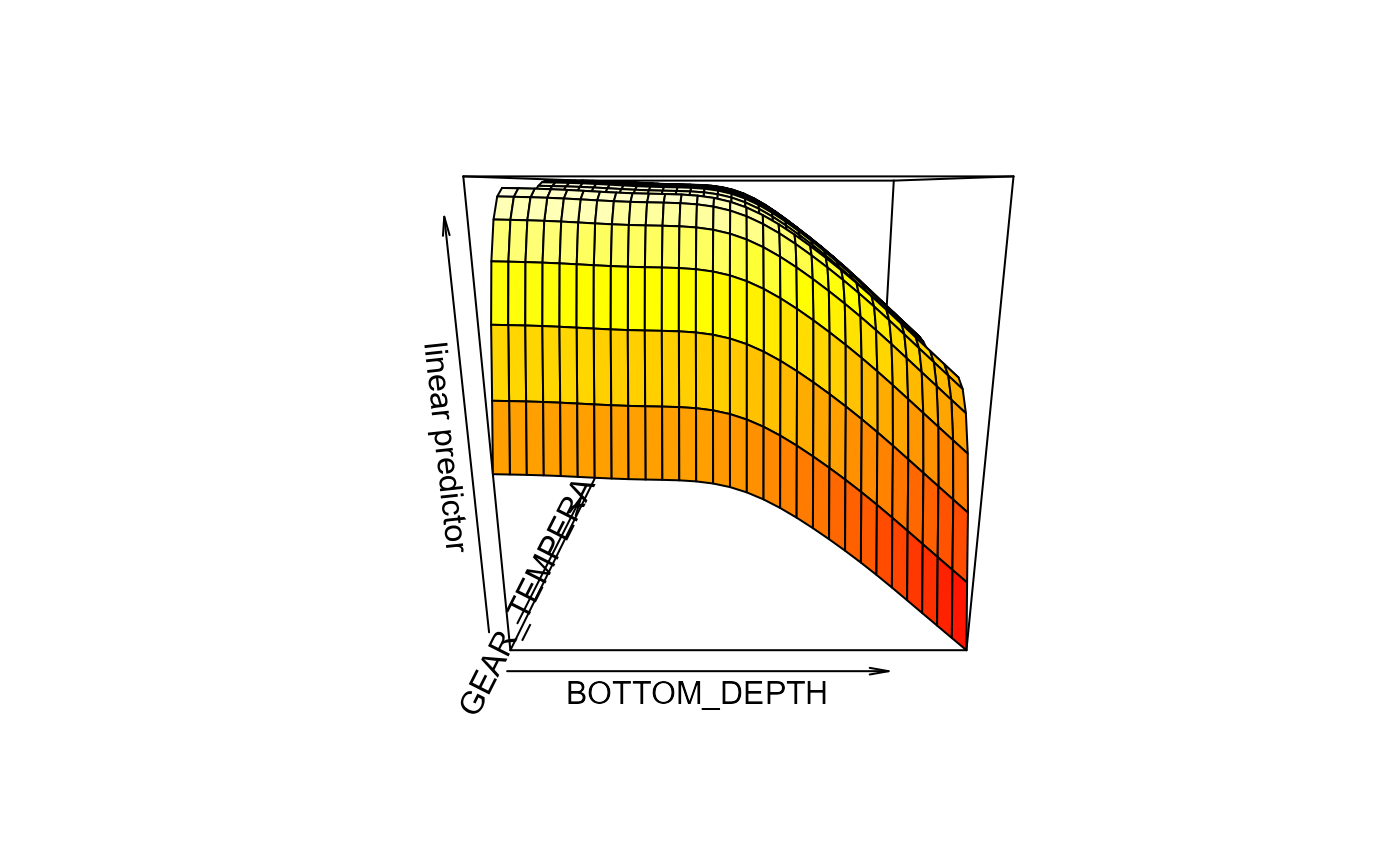
cc <- predict.gam(object = bb, newdata = dat[dat$Year == 2021,])
dat0 <- dat[dat$Year == 2021,] %>%
dplyr::mutate(pred = cc)
figure <- ggplot() +
geom_point(data = dat0,
mapping = aes(x= longitude_dd_start, y = latitude_dd_start, color = pred))
figure